Since March 2022, the cunning Omicron mutant strain once again broke people's peaceful lives, the novel coronavirus epidemics broke out all over the country and affected 30 provinces (autonomous regions and municipalities). As an effective means of precise prevention and control of the SARS-CoV-2 epidemic, nucleic acid detection has become a normal way of life. "Did you do the nucleic acid test today?" It has also become a daily greeting of people. Speaking of which, do you know what core raw materials are needed in nucleic acid detection? This article will introduce the critical core raw material in nucleic acid detection enzyme.
1. Nucleic acid detection process for SARS-CoV-2
2. Core enzymes in nucleic acid extraction
3. Core enzymes during RT-qPCR
4. Core Enzymes of SARS-CoV-2 Nucleic Acid Detection from Yeasen
1. Nucleic acid detection process for SARS-CoV-2
An enzyme is an extremely important class of biocatalysts with high catalytic efficiency and reaction specificity. Most biochemical reactions require the participation of enzymes. In the process of nucleic acid detection of 2019-nCoV (shown in Figure 1), different types of molecular enzymes play an important role in different experimental stages such as nucleic acid extraction and RT-qPCR. Next, according to the different experimental links in nucleic acid detection, we will sort out the core enzyme raw materials used in the nucleic acid detection process.
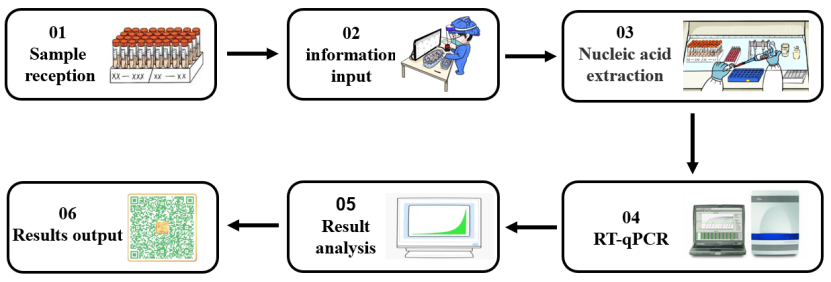
Figure 1. Nucleic acid detection process for SARS-CoV-2
2. Core enzymes in nucleic acid extraction
The novel coronavirus nucleic acid extraction process mainly includes two steps: lysis and purification. Lysis is the process of destroying the cell structure of the sample so that the nucleic acid in the sample is free in the lysis system; Purification is the complete separation of nucleic acid from other components in the lysis system, such as protein, salt, and other impurities, and the reaction process requires the participation of proteinase K, deoxyribonuclease I and RNase inhibitors.
2.1 Protease K
Proteinase K is a serine protease with broad cleavage activity, the cleavage sites are the carboxy-terminal peptide bonds of aliphatic and aromatic amino acids (Figure 2). In the process of nucleic acid extraction, proteinase K can degrade histones which tightly bound with nucleic acids, promote the separation of nucleic acids, and make sample nucleic acid easier to extract. In addition, proteinase K can degrade RNA hydrolase (RNase) activity and inhibit RNase hydrolysis of template RNA.
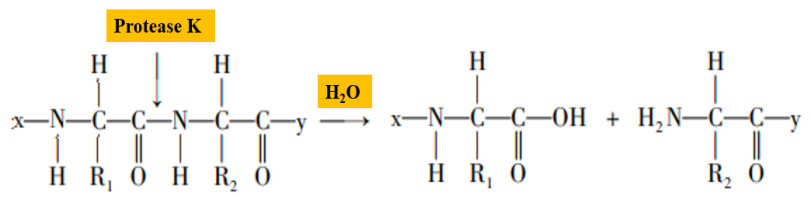
Figure 2. Schematic diagram of proteinase K hydrolyzing peptide bonds
Yeasen Biotech Proteinase K (Cat#10401ES) is derived from recombinant yeast strain, specific activity ≥30 U/mg, free of RNase and DNase, and stable enzyme activity in urea and SDS solution. It is active in a wide pH range (pH 4.0~12.0), suitable for cleavage and removal of proteins such as in situ hybridization sample preparation and nucleic acid purification.
2.2 Deoxyribonuclease I
Deoxyribonuclease I (DNase I) can catalyze various forms of DNA, target to cleavage of phosphodiester bonds adjacent to pyrimidines, and generate polynucleotides with a phosphate group at the 5' end and a hydroxyl group at the 3 ' end, the average digestion product is the smallest polytetranucleotide. In the process of SARS-CoV-2 nucleic acid extraction, DNase I is mainly used to remove genomic contamination in RNA samples, avoid DNA residues in RNA templates and improve the purity of templates.
Yeasen Biotech DNase I (Cat#10325ES) is derived from recombinant E. coli strains, RNase-free, and can be used for the treatment of various RNA samples. the optimal working pH range is 7.0-8.0. In the presence of Mg2+, DNase I can cleave any site of double-stranded DNA randomly; while in the presence of Mn2+, DNase I can cleave DNA double-stranded at the same site, forming blunt ends or 1-2 nucleotide overhanging sticky ends (shown in Figure 3).
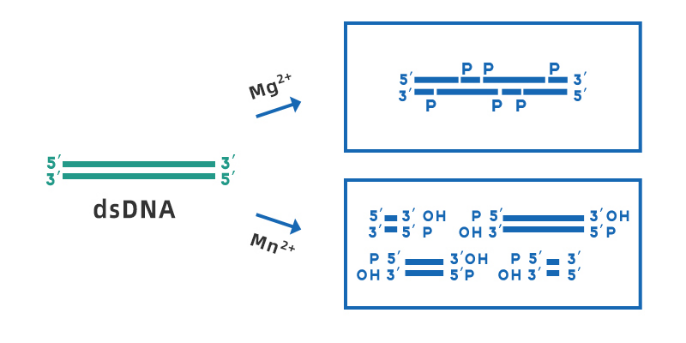
Fig 3. Schematic diagram of DNase I cleaving dsDNA in the presence of Mg2+ and Mn2+
2.3 RNase inhibitor
In the process of SARS-CoV-2 nucleic acid detection, the extraction and purification of sample nucleic acid or the preparation of the experimental reaction system may introduce ribonuclease (RNase) contamination, resulting in the degradation of the RNA template. To avoid RNase contamination, RNase Inhibitor is required.
RNase Inhibitor is a specific RNase inhibitor in human placenta, which can specifically bind RNase to form a complex with a non-covalent bond and inactivate RNase. Yeasen Biotech Murine RNase Inhibitor (Cat#10603ES) does not contain two cysteines that are very sensitive to oxidation in human proteins. It has higher antioxidant activity and can widely inhibit various types of RNases (RNase A, B, C), more suitable for experiments sensitive to high dithiothreitol (DTT) such as qPCR.
3. Core enzymes during RT-qPCR
After the nucleic acid extraction of the SARS-CoV-2 sample is completed, the nucleic acid detection can be completed by RT-qPCR. In the process of these experiments, DNA polymerase, reverse transcriptase, and uracil DNA glycosylase are all essential core enzyme raw materials.
3.1 Reverse transcriptase
After extraction and purification, the SARS-CoV-2 RNA needs reverse transcriptase to catalyze dNTP polymerization to generate a cDNA sequence complementary to the template RNA (Figure 4). For RT-qPCR reaction, high temperature resistant reverse transcriptase should be selected. At present, MMLV reverse transcriptase is the most widely used, because of its lack of DNA endonuclease activity and low RNase H activity, it has more advantages in the application of cDNA cloning.
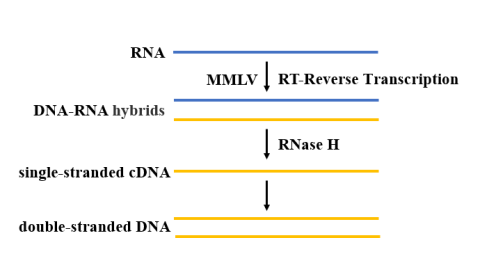
Figure 4. Schematic diagram of the reverse transcription process
Yeasen Biotech Hifair™ V Reverse Transcriptase (Cat#11300ES) is a new reverse transcriptase obtained by genetic engineering technology. It has good thermal stability and can withstand reaction temperatures up to 60°C. And it’s also suitable for reverse transcription of RNA templates with complex secondary structures. At the same time, the enzyme enhances the affinity with the template, is suitable for the reverse transcription of a small number of templates and low-copy genes, and can amplify cDNA up to 10 kb.
3.2 DNA polymerase
After the template reverse transcription process is completed to generate double-stranded cDNA, the "soul player" DNA polymerase in the PCR reaction is required to make an appearance, by polymerizing free deoxyribonucleotides to extend the DNA chain, and a large amount of template DNA are amplified in vitro to achieve the purpose of viral nucleic acid detection.
The DNA polymerase commonly used in RT-qPCR reactions is hot-start Taq DNA polymerase. This type of enzyme is inactive at room temperature. It has polymerization activity only after hot-starting, which can minimize the generation of background signals. It solves the problems of non-specific amplification caused by primer-dimer generation or mismatch in conventional PCR reactions. At present, the commonly used DNA polymerase hot-start modification methods mainly include chemical modification, ligand modification and antibody modification. The principles of different hot-start modification methods are shown in Figure 5.
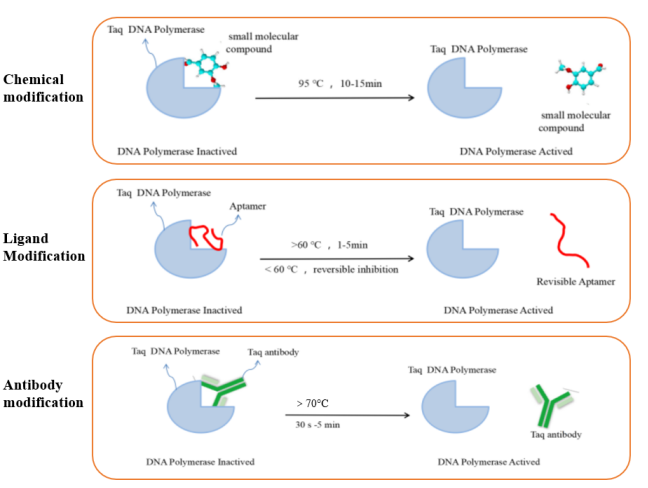
Figure 5. Schematic diagram of different types of modified hot-start enzymes
Yeasen Biotech UNICONTM Hotstart High Specific Taq DNA Polymerase, 5 U/μL (Cat#10726ES) is a double-blocking hot-start DNA polymerase with high template affinity. At room temperature, not only the 5'→3' polymerase activity is blocked, but also the 5'→3' exonuclease activity is blocked. Denaturation at 95°C for 30 sec can completely inactivate the blocking antibody, releasing DNA polymerase activity and exonuclease activity. The double-blocking feature can not only effectively prevent non-specific amplification caused by mismatches or primer-dimers, but also effectively inhibit the decrease of fluorescence signal caused by probe degradation. The double guarantee makes the in vitro detection reagents more stable during transportation or uses at room temperature.
3.3 Uracil DNA glycosylase
In the process of new coronavirus nucleic acid detection, aerosol pollution in the operating environment is the most common factor causing false positive PCR results. Adding UDG enzyme (Uracil DNA Glycosylase, uracil DNA glycosylase) to the amplification system can effectively eliminate the amplification residual pollutants (mostly in the form of aerosols) mixed in the PCR system to ensure the accuracy of the amplification results. The anti-pollution principle of the UDG enzyme is shown in Figure 6.
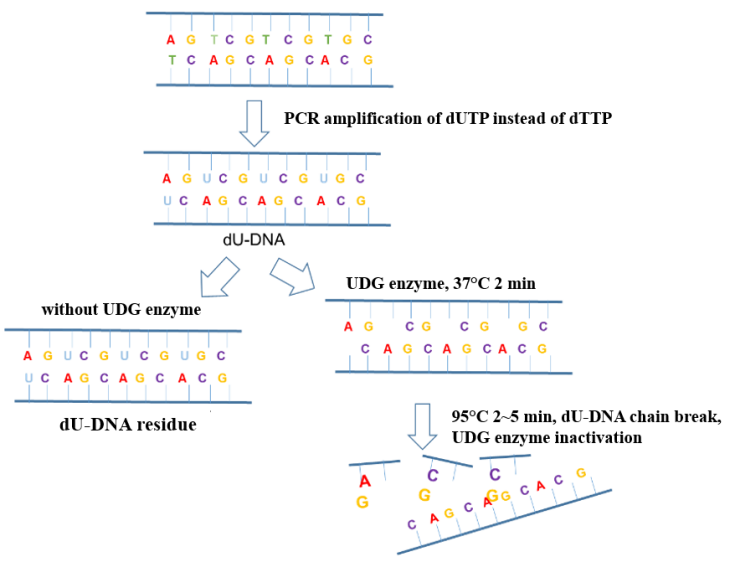
Figure 6. Schematic diagram of the anti-pollution principle of UDG enzyme
Yeasen Biotech Uracil DNA Glycosylase (UDG/UNG), heat-labile,1 U/μL (Cat#10303ES) is active at 25-37°C, heat-sensitive, and irreversibly inactivated at 50°C for 10 min or 95°C for 2 min. There is no endonuclease and RNase residue, and the host bacteria genome residue is less than 10 copies. It can be used in conjunction with hot-start Taq DNA polymerase to ensure the specificity of the amplification reaction.
4. Core Enzymes of SARS-CoV-2 Nucleic Acid Detection from Yeasen
|
Process |
Description |
Product name |
SKU |
|
Sample processing |
Protein digestion |
10401ES |
|
|
RNA extraction |
Recombinant DNase I (RNase-free) (Inquire) |
10325ES |
|
|
RNase inhibition |
10603ES |
||
|
Reverse transcription |
Suitable for RT-qPCR |
11300ES |
|
|
HifairTM V Reverse Transcriptase (600U/ μL) GLyceroL- free (Inquire) |
11301ES |
||
|
PCR amplification |
Hot-start DNA polymerase |
Hieff UNICONTM Hotstart High Specific Taq DNA Polymerase, 5 U/μL |
10726ES |
|
Thermal UDG |
10303ES |
Regarding reading:
Reverse Transcriptase Selection
YEASEN Heat-labile UDG——Easily control aerosol pollution
Murine RNase Inhibitors-Successfully eliminate RNase contamination and preserve RNA