Minimalist Transcriptional Assembly Library Kit, Quickly Play RNA-seq
Transcriptome in a broad sense refers to the collection of all transcription products in a cell under a certain physiological condition, including messenger RNA, ribosomal RNA, transfer RNA, and non-coding RNA; in a narrow sense, it refers to the collection of all mRNAs. Through high-throughput sequencing of the mRNA of a certain species, the overall transcriptional activity of a specific species can be detected at the single nucleotide level, so that almost all transcript information of the species in a certain state can be obtained comprehensively and rapidly.
2. RNA library construction method recommendation
1. mRNA enrichment methods
Because transcriptome sequencing can obtain an abundance of information on all RNA transcripts, and its high accuracy, it has a wide range of applications. Transcriptome sequencing can be divided into the eukaryotic transcriptome and prokaryotic transcriptome according to the species of the sample to be tested. The structure of eukaryotic and prokaryotic mRNA is different, so the methods of pre-enrichment are also inconsistent.
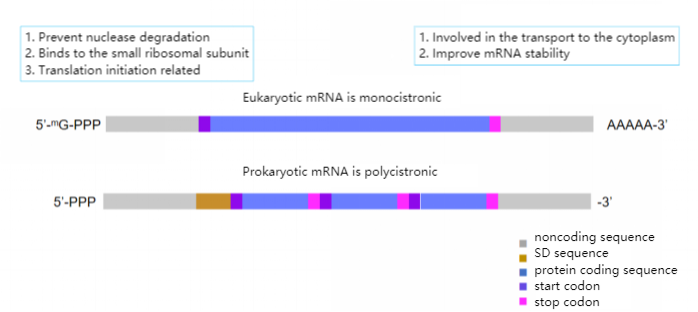
Figure 1. Difference between eukaryotic and prokaryotic mRNA
Eukaryotic mRNA contains a poly-A tail structure, so oligo-T modified magnetic beads can be used for specific enrichment of mRNA; however, prokaryotic mRNA does not have a poly-A structure like eukaryotic mRNA, so it cannot be The mRNA is directly purified by oligo-T magnetic beads, so prokaryotes generally can enrich the target RNA by the reverse enrichment method, that is, removing the rRNA in the Total RNA. Then, through the steps of fragmentation, reverse transcription one-strand synthesis, two-strand synthesis, adding A at the end, adapter ligation, and library amplification, a library that can be sequenced on the computer is formed.
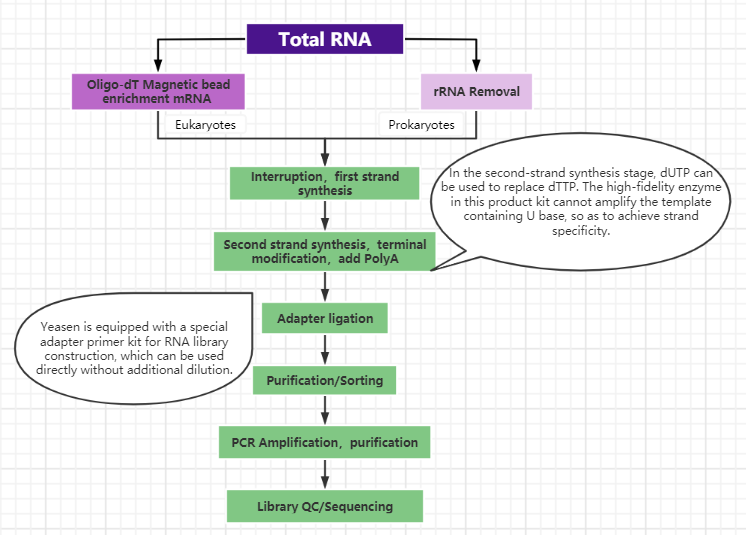
Figure 2. Yeasen provides different mRNA enrichment methods for library-building solutions
2. RNA library construction method recommendation
Most of the traditional RNA library construction methods are complicated and time-consuming, and there are also many technical challenges in the detection of trace samples. Coupled with the high frequency of infectious diseases in recent years, the rapid processing of RNA-based unstable samples is particularly important. Hieff NGS™ Ultima Dual-mode RNA Library Prep Kit is a high-efficiency RNA library construction kit. Through process optimization, the steps of double-strand synthesis and the final addition of A are combined into one step, which greatly reduces the experimental operation time. At the same time, users can Choose RNA enrichment modules for library construction.
3. Popular science tips
Since the mRNA is reverse transcribed into double-stranded cDNA during the construction of the transcriptome library, during sequencing, the sequence information of the double-stranded cDNA will be obtained at the same time, which makes it impossible to distinguish the sense and antisense strands in the cDNA. Strand-specific libraries, on the other hand, retain mRNA orientation information during library preparation and sequencing, resulting in more effective data and more accurate gene expression quantification; more accurate gene annotation, and more reliable analysis of new gene identification; the same method can identify and open Reading frame antisense ncRNA, found antisense transcript.
3.1 Performance Demonstration 1 - rRNA Removal Reverse Enrichment
Sample Information: Different DV200 Human FFPE RNAs
Kit: Yeasen: Hieff NGS™ Ultima Dual-mode RNA Library Prep Kit (Cat#12252)
Competing product: N* brand
Other reagents:
rRNA Depletion: Hieff NGS™ MaxUp Human rRNA Depletion Kit (rRNA & ITS/ETS) (Cat#12257)
RNA purification: Hieff NGS™ RNA Cleaner (Cat#12602)
DNA Purification: Hieff NGS™ DNA Selection Beads (Cat#12601)
Library Quantitation: dsDNA HS Assay Kit for Qubit (Cat#12640)
Library Adapter: Hieff NGS™ RNA 384 CDI Primer for Illumina (Cat#12414)
3.1.1 Sequencing data display
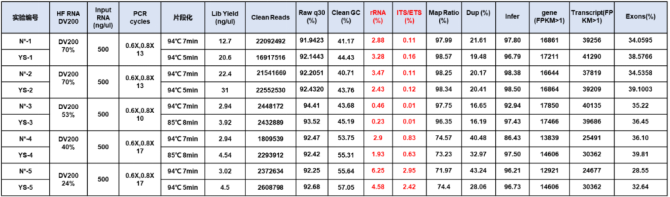
Figure 3. Sequencing data display
*DV200 represents the proportion of RNA fragments larger than 200nt in the sample. For severely degraded FFPE samples, the DV200 value can better reflect the quality of the sample.
3.1.2 Consistency comparative analysis
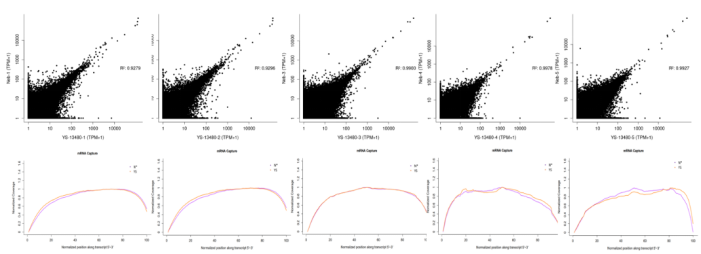
Figure 4. Consistency comparative analysis
3.2 Performance display 2 - Oligo-dT magnetic bead enrichment
Sample Information: Model Crop Arabidopsis RNA
Kit: Yeasen: Hieff NGS™ Ultima Dual-mode RNA Library Prep Kit (Cat#12252)
Competing products: N* brand, V* brand
Other reagents:
mRNA purification: Hieff NGS™ mRNA Isolation Master Kit (Cat#12603)
DNA Purification: Hieff NGS™ DNA Selection Beads (Cat#12601)
Library Quantitation: dsDNA HS Assay Kit for Qubit® (Cat#12640)
Library Adapter: Hieff NGS™ RNA 384 CDI Primer for Illumina® (Cat#12414)
3.2.1 Sequencing data display
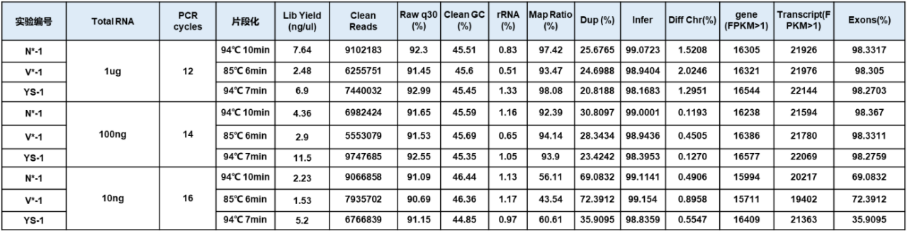
Figure 5. Sequencing data display
3.2.2 Consistency comparative analysis
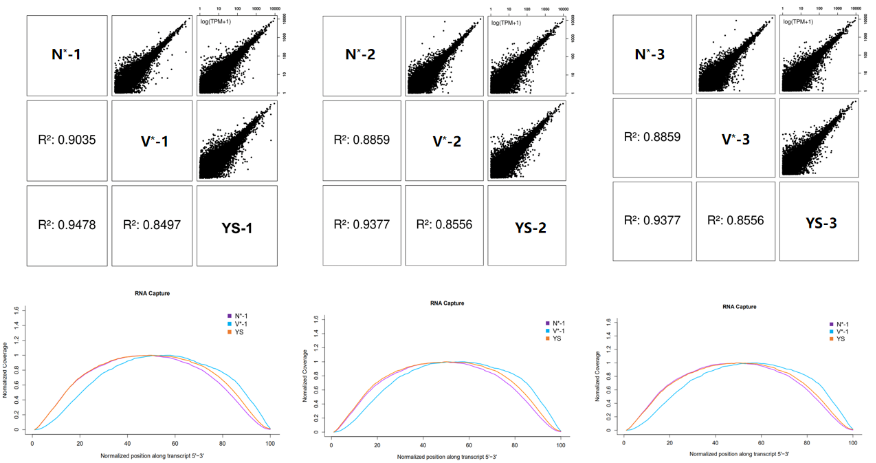
Figure 6. Consistency comparative analysis
4. Product information
The related products that Yeasen can provide are shown in Table 1.
Table 1. Product information
| Classification | Product Name | SKU | Specifications | |
| RNA-seq | Hieff NGS™ MaxUp Human rRNA Depletion Kit (rRNA & ITS/ETS) MaxUp | 12257ES | 12257ES08 | 8T |
| 12257ES24 | 24T | |||
| 12257ES96 | 96T | |||
| Hieff NGS™ Ultima Dual-mode RNA Library Prep Kit | 12308ES | 12308ES08 | 8T | |
| 12308ES24 | 24T | |||
| 12308ES96 | 96T | |||
| Hieff NGS™ Ultima Dual-mode mRNA Library Prep Kit | 12309ES | 12309ES08 | 8T | |
| 12309ES24 | 24T | |||
| 12309ES96 | 96T | |||
5. Regarding reading
Various types of magnetic beads in NGS: DNA\RNA\mRNA magnetic beads
The solution for the NGS library preparation from DNA samples