5 minutes to understand the past and present of DNA selection beads (including result analysis and FAQ interpretation)
DNA selection beads are an essential product in the high-throughput sequencing process. Through the principle that the active groups of magnetic particles can bind and dissociate with nucleic acids under certain conditions, the target fragments in the sample are separated. It can realize high-throughput automated operation of nucleic acid samples, and is widely used in the fields of gene sequencing and molecular diagnosis.
Background
The idea for the invention of DNA selection beads originally came from chemist John Ugelstad of the Norwegian University of Science and Technology, who in 1976 used polystyrene as the main material to produce uniformly magnetized spherical particles. In 1979, Vogelstein et al. reported that in the presence of high concentrations of sodium iodide, glass powder was used as an adsorbent to extract DNA fragments from agarose gels, and then based on silica gel and other solid-phase nucleic acid purification techniques with hydrophilic surface carriers. widely developed. The nucleic acid purification method based on magnetic particles is one of them.
Structure
Although the nano-scale DNA selection beads are various so far, with different surface properties and different separation principles, their material composition and basic structure are not much different. The basic structure is divided into three layers, the innermost layer is polystyrene, the second layer is wrapped with magnetic substances (usually Fe3O4), and the outermost layer is modified functional groups (such as carboxyl groups). Among them, the functional groups can be combined with nucleic acids, and the surface groups are different, and the downstream applications are also different, such as nucleic acid extraction, purification, biotin capture, etc.
Principles
Commercial DNA selection bead product systems generally include: DNA selection beads, polyethylene glycol (PEG), salt ions, etc. Based on SPRI (Solid Phase Reversible Immobilization) technology, in the presence of a certain concentration of PEG and under the promotion of NaCl or MgCl2, the DNA molecular conformation will change dramatically, exposing a large number of negatively charged phosphate groups on the phosphate backbone. , bound to the negatively charged surface carboxyl DNA selection beads. It is currently believed that this interaction between negative and negative charges is due to the effect of positively charged salt ions (such as Na+). The negatively charged phosphate group forms an ion bridge with the carboxyl group through the dissociated salt ion (such as Na+), so that the DNA is specifically adsorbed to the surface of the carboxyl DNA selection bead. Using the magnetic properties of DNA selection beads, collection and elution can be performed by an external magnetic field.
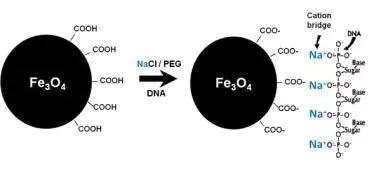
Figure: Schematic diagram of the principle of DNA adsorption by DNA selection beads
Operation
Benefiting from the development of high-throughput sequencing technology, DNA selection beads are also highly regarded for their high-throughput and automation features, and are widely used in DNA purification/double-round sorting (chip screening) in NGS library construction.
The purpose of DNA purification is to remove unwanted fragments. Since DNA selection beads preferentially adsorb large fragments, a certain proportion of DNA selection beads can be used to adsorb fragments larger than a certain size, and the supernatant (removal of non-target fragments) can be discarded and the DNA adsorbed by DNA selection beads can be removed. Elution recovery. Commonly used for the removal of small fragments such as adapter dimers/primer dimers.
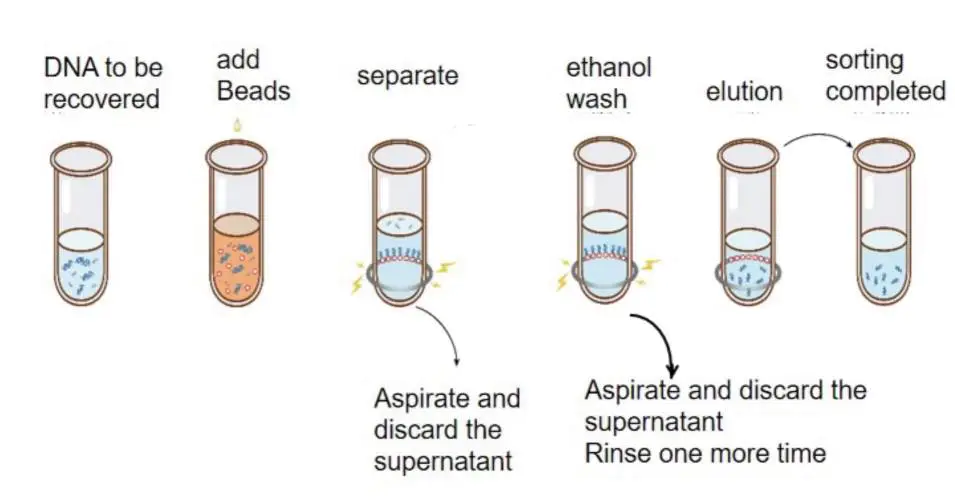
Figure: Schematic diagram of DNA purification operation
The purpose of double-round DNA sorting (slice screening) is to select the size of DNA fragments in specific regions from fragmented DNA samples. Since DNA selection beads preferentially adsorb large fragments, two rounds of DNA selection bead manipulation are required for sorting. Take the range of sorting 450-550 bp fragments as an example: First, the DNA selection beads adsorb fragments of more than 550 bp in the first round, then discard the DNA selection beads and leave the supernatant, and the fragments in the supernatant are the fragments below 550 bp. In the second round, the DNA selection beads were taken to adsorb the DNA selection beads above 450 bp (below 550 bp). At this time, the supernatant was discarded, and the target fragments on the DNA selection beads were eluted and recovered. Commonly used for NGS library construction fragment sorting (chip screen).
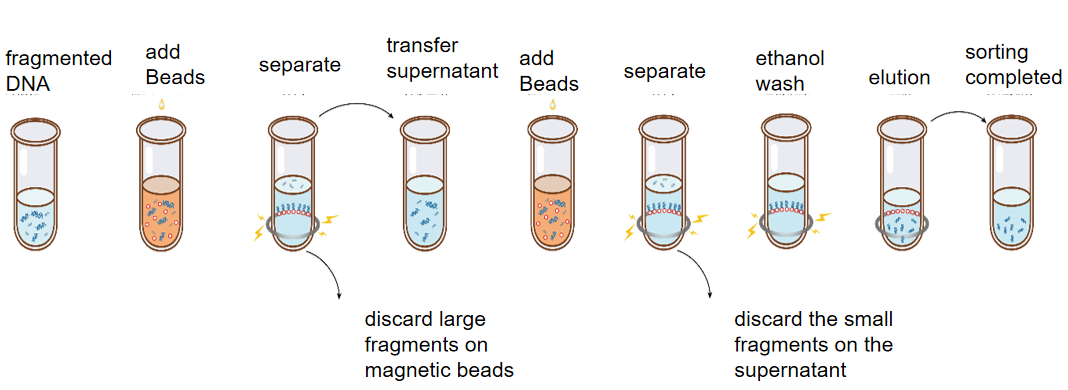
Figure: Schematic diagram of DNA double-round sorting operation
Result analysis
1. DNA selection bead recovery efficiency
Part of the DNA sample will be lost after being purified by DNA selection beads, and the recovery efficiency of DNA selection beads can be used to evaluate the recovery ability of DNA selection beads. Specifically, it can be calculated by: recovery efficiency = (purified DNA quality/Input DNA quality) × 100%.
Take the following table as an example, under the same ratio conditions: the recovery efficiency of test A sample of test person 1 = 153.25/169.5 = 90.41%, and the recovery efficiency of other samples can be calculated in the same way.
| Test people | Sample number | Input(ng) | Total after purification | Recycling efficiency(%) | Average recycling efficiency(%) |
|---|---|---|---|---|---|
| Test people 1 (1.8x) |
A | 169.5 | 153.25 | 90.41 | 90.41 |
| B | 159.75 | 90.25 | 94.5 | ||
| C | 162 | 95.58 | |||
| D | 158.75 | 93.66 | |||
| Test people 2 (1.8x) |
A | 169.5 | 154.5 | 91.15 | 91.15 |
| B | 155 | 91.45 | 91.94 | ||
| C | 154.25 | 91 | |||
| D | 158.25 | 93.36 |
2. DNA selection bead sorting accuracy
Due to the limitation of short read length of next-generation sequencing segments, the final NGS library needs to meet certain length requirements. Therefore, fragments of a specific length range may be sorted in the NGS library building to meet the requirements of the computer. The sorting accuracy is to evaluate the size of the fragments sorted by different DNA selection bead input ratios. It is generally detected with an Agilent 2100 instrument.
Under the condition of a specific DNA selection bead ratio (0.45×/0.15×), the distribution ranges of fragments after sorting of different samples are concentrated in the same position. A better sorting effect should be a single peak with a narrow and rounded top (as shown in the figure below).
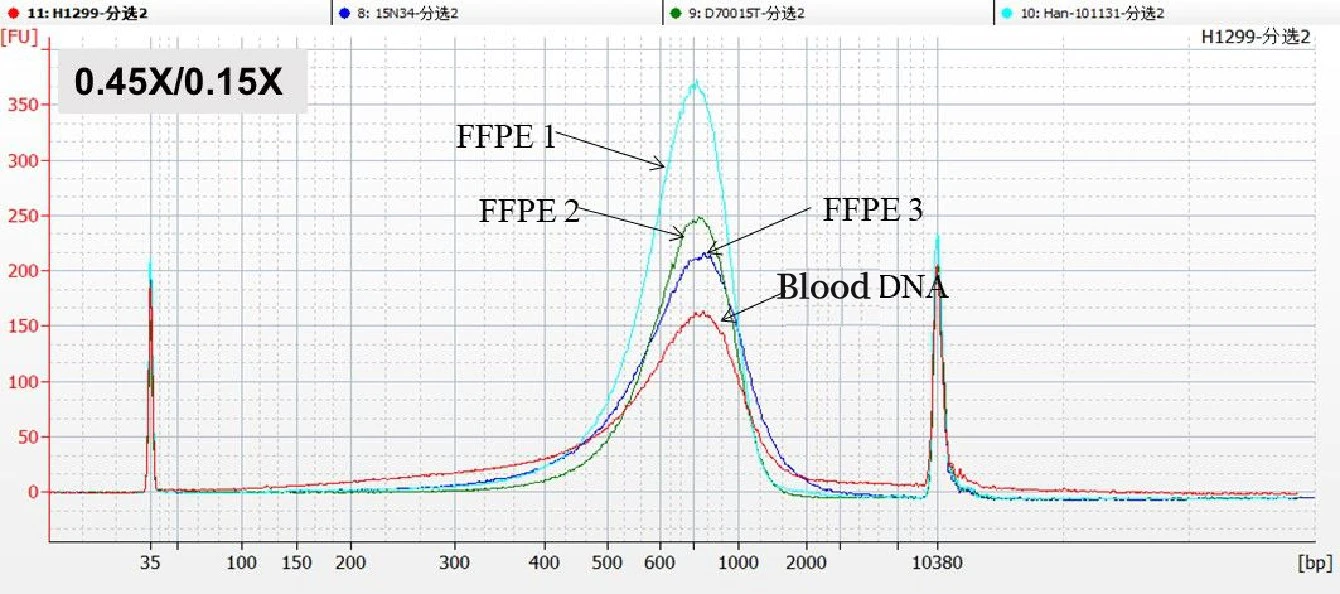
【Note】: The data results come from a biological company in Shanghai. It is worth noting that the different DNA selection bead buffers of different manufacturers lead to inconsistent range of fragments obtained under the same ratio. Please select the appropriate separation ratio according to the corresponding DNA selection bead instructions before use.
Precautions
1. The DNA selection beads need to be equilibrated to room temperature before use, otherwise the experimental effect will be affected (PEG separation effect is easily affected by pH, temperature, etc.);
2. The 80% absolute ethanol used for DNA selection bead elution should be prepared immediately;
3. It is recommended that the initial volume be ≥100 μL for length sorting. If it is insufficient, please use ultrapure water to make up the volume (if the sample volume is too small, the pipetting error will increase, which will affect the sorting accuracy);
4. When sorting DNA selection beads, pay special attention to discarding the DNA selection beads containing large fragments after the first round of sorting, and discarding the supernatants containing small fragments in the second round;
5. During the first round of sorting and transferring the supernatant, avoid attracting the DNA selection beads, causing large fragments to remain;
6. If the separation/purification product needs to be stored for a long time, please use TE buffer to elute and store.
FAQs
The seemingly simple purification/sorting experiment has various unexpected situations in the actual experiment. What should we pay attention to in our actual operation in terms of DNA selection bead product selection and product use?
1.Applications
Q1. Can DNA selection beads adsorb ssDNA?
A: Single-stranded DNA can be recovered, but the specific performance has not been tested. The attached figure is the data of single-stranded DNA recovered by Beckman DNA selection beads, which can be used as a reference.
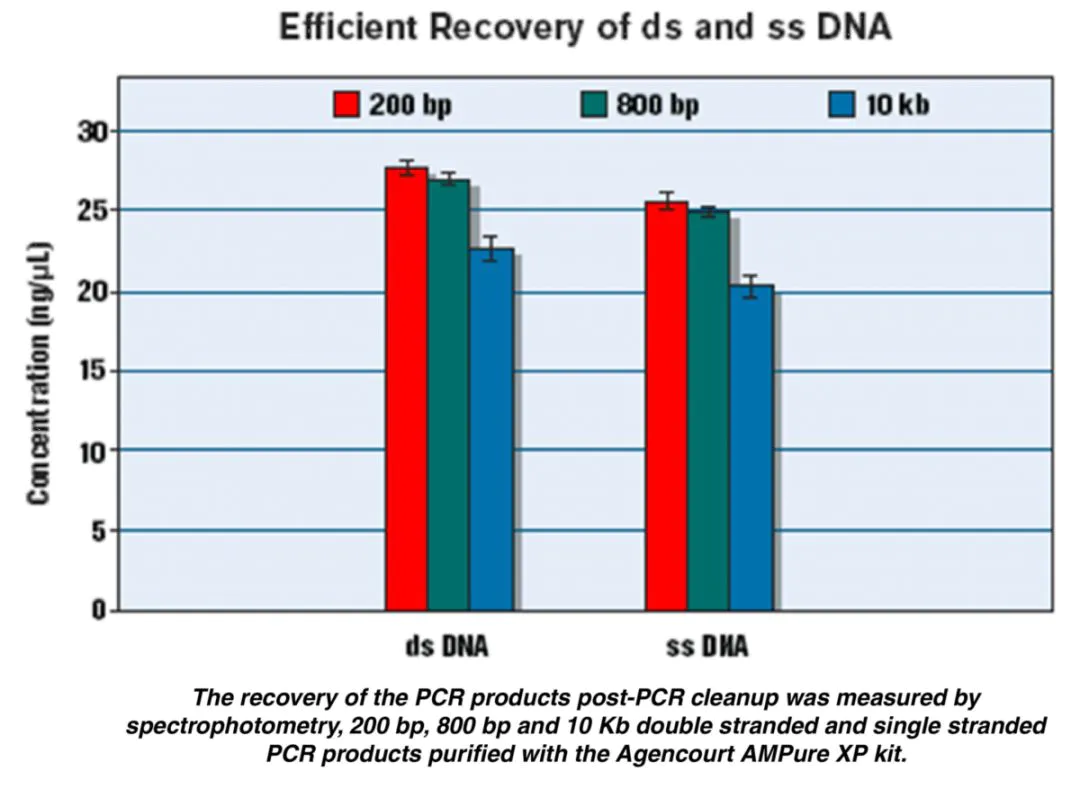
Q2. Can sorting DNA selection beads purify gDNA?
A: Yeasen DNA selection Separation Beads have tested fragments with a maximum size of 20kb, and the recovery rate is >90%. Genomic DNA has not been tested. Due to the tight binding of large DNA to DNA selection beads, it is recommended to increase the volume of eluent as much as possible to avoid excessive drying time.
Q3. How about the DNA binding ability of DNA selection beads?
A: In the current DNA selection bead system, the adsorption capacity of DNA selection beads is supersaturated, and customers do not need to worry about the insufficient binding capacity of DNA selection beads. According to known reports, 7 μg of DNA can be bound per 1 μL of DNA selection beads.
Q4. When using DNA selection beads to adsorb small fragments, will large fragments also be adsorbed?
A: DNA selection beads preferentially adsorb large fragments. When the input amount of DNA selection beads is increased, the DNA selection beads will enhance the ability to adsorb small fragments on the basis of adsorbing large fragments. for example:
Referring to the DNA selection bead purification and sorting table, add 80 μL of DNA selection beads Cat#12601 to 100 μL of sample. At this time, fragments >350 bp are adsorbed on the DNA selection beads. However, when the amount of DNA selection beads added reaches 100 μL, the Fragments >200 bp were adsorbed.
2.Product use effect
Q1. After sorting/purifying with DNA selection beads, what is the matter with small fragments remaining?
A: Most of the small fragment residues are adapter dimers/primer dimers. If purifying, reduce the proportion of DNA selection beads; if sorting: appropriately reduce the proportion of DNA selection beads in the second round, which can effectively improve this kind of situation.
Q2. What are the possible reasons for the low bead recovery?
A: 1) The DNA selection beads and DNA were not mixed well enough to be fully adsorbed. It is recommended that the reaction system be thoroughly mixed;
2) If the ratio of DNA selection beads is wrong, it is recommended to select the appropriate ratio according to the instructions;
3) Insufficient incubation time, ensure that the incubation time is at least 5 min;
4) During DNA selection bead sorting, the DNA selection beads are attracted during the second round of DNA selection bead adsorption. A 10 μL pipette tip can be connected in series in front of the 200 μL pipette tip for pipetting to prevent DNA selection beads from being attracted;
5) The ethanol used for washing DNA selection beads is not prepared, resulting in too low ethanol concentration. It is recommended that the ethanol concentration should not be lower than 70%;
6) Excessive drying: the DNA selection beads are dried for a long time, resulting in surface cracks, DNA is difficult to elute, and the yield is reduced; it is recommended that the drying time should not be too long, and the surface should just be cracked;
7) Insufficient eluent volume: The final eluent should completely cover the DNA selection beads in the tube.
3.product storage
Q: The DNA selection beads were accidentally frozen at a low temperature (-20°C) for a period of time, can they still be used?
A: It cannot be used. Low temperature will destroy the modification of the surface group of the DNA selection beads and affect the recovery performance of the DNA selection beads.
About Us
Yeasen Biotechnology is a high-tech enterprise focusing on innovative research and development of molecular enzyme raw materials. DNA selection beads are an essential product for NGS library construction. According to the market demand for DNA selection beads, YEASEN concentrates professionals and is committed to the innovative development of DNA selection bead series products. At present, it has a variety of nucleic acid purification DNA selection bead products, which can be applied to high-throughput sequencing and nucleic acid purification.
Product information
| Product name | SKU | Specifications | |
| Hieff NGS® DNA selection Beads(Superior Ampure XP alternative) | 12601 | 12601ES03 | 1 mL |
| 12601ES08 | 5 mL | ||
| 12601ES56 | 60 mL | ||
| 12601ES75 | 450 mL | ||
| Hieff NGS® Smarter DNA Clean Beads | 12600 | 12600ES03 | 1 mL |
| 12600ES08 | 5 mL | ||
| 12600ES56 | 60 mL | ||
| 12600ES75 | 450 mL | ||