GMP-grade reagents for mRNA in vitro synthesis
mRNA vaccines can express target proteins in the human body. Compared with traditional small molecule drugs and antibody drugs, mRNA vaccines are more likely to act on intracellular targets. With the rapid development of biotechnology, mRNA vaccines based on in vitro RNA synthesis technology have had a huge impact on COVID-19 in 2020. How does the mRNA vaccine work? And what is the process and focus of mRNA vaccine manufacturing? Please continue reading.
1. How does the mRNA vaccine work?
2. How is mRNA synthesized in vitro?
3. Key factors affecting the quality of mRNA vaccines
4. Products related to mRNA vaccine production
1. How does the mRNA vaccine work?
mRNA is a type of single-stranded ribonucleic acid, which is transcribed for a strand of DNA as a template. It carries genetic information and can guide protein synthesis, which best describes the role of mRNA in protein synthesis. mRNA teaches the body to make a protein that looks like a virus but is harmless. The body responds to this protein to produce cells and antibodies that recognize and fight against the virus. This is related to the function of mRNA in protein synthesis. The mRNA vaccine needs to go through the following steps:
First, the mRNA is wrapped by various carriers and injected into the human body. Then, the liposome wrapping the mRNA enters the cell through endocytosis, releasing the mRNA in the cell. Finally, the human organelle is used to translate and express the antigenic protein, stimulating the body to produce an immune response. The effect of the mRNA vaccine is closely related to mRNA construct and delivery. Two categories of mRNA constructs are currently being actively evaluated, namely non-replicating mRNA (NRM) constructs and self-amplifying mRNA (SAM) constructs.
As figure 1 shows, NRM and SAM can deliver the transcript of interest, encoding one or more immunogen(s), into the host cell cytoplasm where expression generates translated protein(s) to be within the membrane, secreted or intracellularly located. NRM and SAM are prepared in the form of lipid nanoparticles (LNPs) to avoid degradation by cells and promote cellular uptake. They enter the cytoplasm through the endocytic pathway to the cell membrane. The NRM constructs are immediately translated by the ribosomes to produce the protein of interest, while SAM is ribosomal translation produces the replicate necessary for mRNA amplification, and the self-amplifying mRNA construct is translated by the ribosome to produce the protein of interest, which undergoes subsequent post-translational modification.
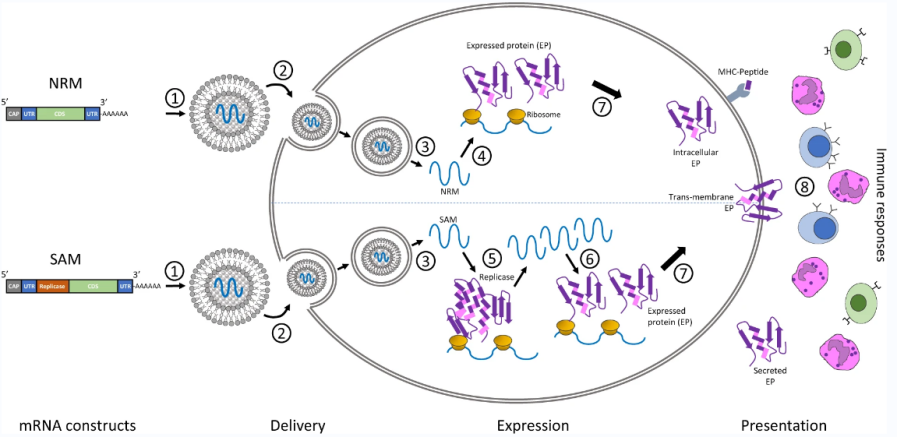
Figure 1: mRNA constructs and delivery technologies[1]
2. How is mRNA synthesized in vitro?
The mRNA vaccine production process includes identification of target pathogens, genome sequencing, electron transfer of the sequence, vaccine design, pilot mRNA vaccine production, and validation, GMP manufacturing of mRNA vaccines, and vaccination.
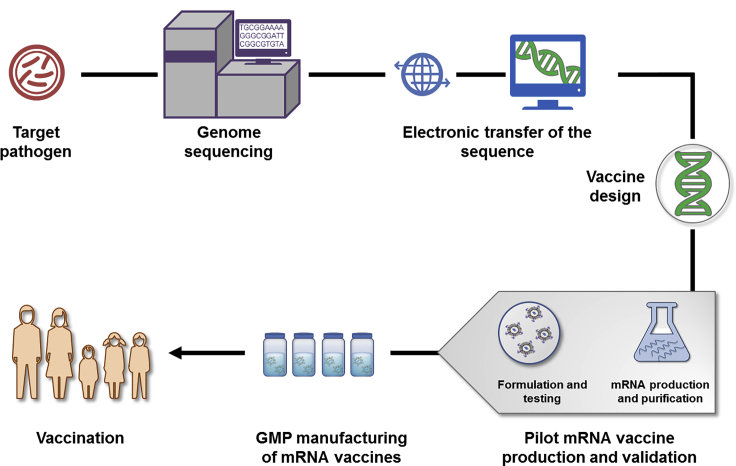
Figure 2: Schematic Illustration of mRNA Vaccine Production[2]
The in vitro synthesis of mRNA is an important step in the production of mRNA vaccines. It refers to the use of DNA as a template for a cell-free system in vitro to complete mRNA synthesis by RNA polymerase, and then add partial modifications to it to simulate in vivo translation process.
The main steps are plasmid linearization, mRNA transcription, DNA template removal, RNA purification, and capping reactions. The mRNA synthesized in vitro is different from the endogenous cellular mRNA. After transfection into the cell, it can be translated immediately and efficiently to express the target protein without the process of RNA shearing and nuclear export. The construction of optimally translated IVT mRNA suitable for therapeutic use has been reviewed previously. Briefly, IVT mRNA is produced from a linear DNA template using a T7, a T3, or a Sp6 phage RNA polymerase. The resulting product should optimally contain an open reading frame that encodes the protein of interest, flanking UTRs, a 5' cap, and a poly(A) tail. The mRNA is thus engineered to resemble fully processed mature mRNA molecules as they occur naturally in the cytoplasm of eukaryotic cells.
3. Key factors affecting the quality of mRNA vaccines
3.1 Purity
The purity of the mRNA is a crucial determinant of yields, and it is known to produce oligoribonucleotide but also type I interferon and inflammatory cytokine. Karikó et al. demonstrate that the removal of contaminants in mRNA preparations reduced innate immune responses and resulted in significantly higher levels of reporter protein expression in vitro.
3.2 Sequence selection
Sequence selection is very important to correctly select the antigen sequence of the virus, which is the direction of mRNA vaccine development because different proteins are encoded by different RNA sequences. The host can synthesize any protein based on the encoded information carried by the mRNA, so the selection of antigens for mRNA vaccines is very wide, but the correct antigen selection of a specific virus must be very careful, and it is necessary to ensure that the vaccine containing the RNA is both safe and effective.
3.3 Chemical modification
The main purpose of chemical modification is to enhance mRNA stability, and translation efficiency and reduce immunogenicity. The necessary elements for the structural composition of mRNA include the Cap, the 5' UTR region, the 3' UTR region, the ORF encoding the antigenic protein, and the Poly (A) tail structure.
3.3.1 The Cap structure
The Cap structure is a necessary structure of translation initiation, providing a signal for ribosomes to recognize mRNA. And Cap acts as a self-recognition signal to avoid immunosuppression caused by activation of Rig-I and IFIT.
3.3.2 The 5' UTR region
The structural features of the 5' UTR region are one of the main factors that affect the efficiency of mRNA translation. In eukaryotic cells, mRNA needs to recruit ribosomal subunits to bind to the 5'm7G cap before translation. As the start codon is usually located far downstream of the 5'm7G cap, so the ribosomal subunit needs to go through the 5' UTR to reach the start codon and start the translation. The length and structure of the 5' UTR, therefore, have a significant impact on the initiation of translation.
3.3.3 The ORF of mRNA
In the ORF of mRNA, codon optimization refers to the replacement of commonly used codons with less commonly used codons, a process that can improve mRNA stability and translation efficiency. When mRNA is injected into the human body, although the codons contained in the mRNA of the original host correspond to the same amino acids, they are not commonly used, resulting in the instability of mRNAs after entering the human body, and the translation efficiency is low, so codons need to be optimized.
3.3.4 The 3′ UTR region
The 3′ UTR region is an important factor of mRNA instability, among which AU-rich sequences (AREs), AUUUA repeats, and GU-rich sequences (GREs) are the most common causes of 3′ UTR-induced mRNA instability. These sequences should be avoided in the mRNA.
3.3.5 The Poly(A) tailed
Poly(A) tailed is an important factor affecting the translation efficiency and stability of mRNA, and its synergistic effect of 5′Cap can improve translation efficiency, while the removal of Poly(A) tailed is the first step in the degradation of most eukaryotic mRNAs. The rate-limiting step and the Poly(A) tailed inhibit mRNA degradation and recapping.
4. Products related to mRNA vaccine production
4.1 Products related to the process of in vitro synthesis of mRNA
As mentioned in section 2, in vitro synthesis of mRNA requires corresponding reagents. Details are as follows:
Table 1: Products related to the process of in vitro synthesis of mRNA
|
Plasmid linearization |
In vitro transcription |
DNA template removal |
RNA purification |
Capping reaction |
RNA purification |
|
|
|||||
|
|
|||||
4.2. Popular kit used for in vitro transcription
The kits required for in vitro transcription of mRNA can perform in vitro transcription experiments in a relatively short period and synthesize high yields of RNA transcripts. The product features of the T7 High Yield RNA Synthesis Kit are firstly high yielding (100-200μg in 2 hours, and a single amplification reaction can produce microgram-level mRNA), and secondly, versatility (suitable for long and short fragments) RNA transcription), and finally high flexibility (untagged, tagged and capped mRNAs are available). The reagents provided are as follows:
Table 2: Popular kit used for in vitro transcription
|
Contents No. |
Name |
Catalog No./Specification |
|
|
10623ES50 (50 T) |
10623ES60 (100 T) |
||
|
10623-A |
T7 RNA Polymerase Mix |
100 μL |
200 μL |
|
10623-B |
10×Transcription Buffer |
100 μL |
200 μL |
|
10623-C |
ATP(100mM) |
100 μL |
200 μL |
|
10623-D |
CTP(100mM) |
100 μL |
200 μL |
|
10623-E |
GTP(100mM) |
100 μL |
200 μL |
|
10623-F |
UTP(100mM) |
100 μL |
200 μL |
|
10623-G |
Control DNA Template(500ng/μL) |
10 μL |
20 μL |
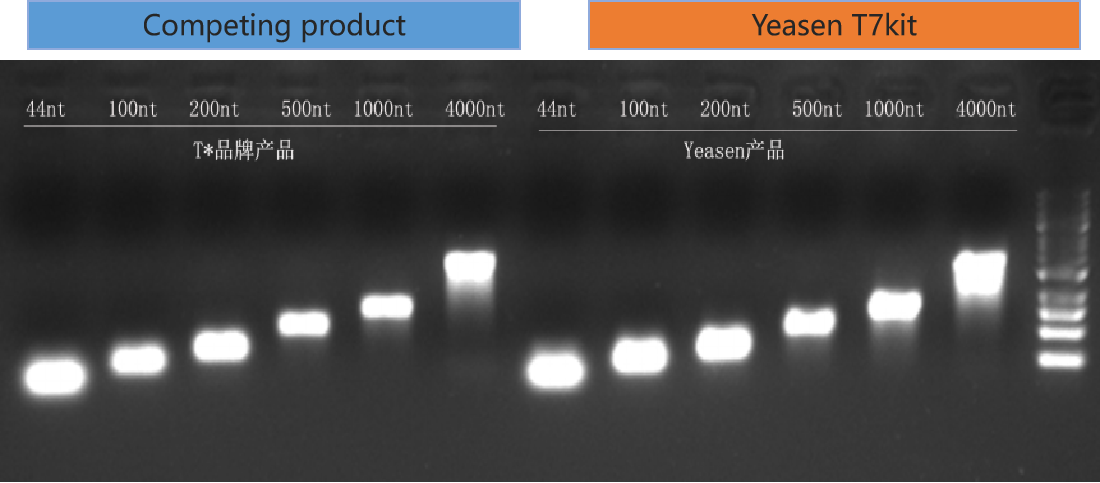
Figure 3. As compared to the competing product, Yeasen T7 kit yield a comparable amount of mRNA
4.3. Popular capping enzyme in mRNA modification
In eukaryotes, mRNA is post-transcriptionally modified to form a special structure of the 5' end, namely the Cap structure, which plays an important role in mRNA stability, transport, and translation. Vaccinia virus capping enzyme is an effective enzyme that catalyzes the formation of a cap structure. It consists of two subunits, D1 and D12. It has both RNA triphosphatase activity, guanyl acyltransferase activity, and guanine methyltransferase activity. A 7-methylguanine cap structure (m7Gppp) is attached to the 5' end of the RNA (m7Gppp5'N). mRNA Vaccinia Capping Enzyme GMP-grade has the characteristics of a high standard, no residue, high purity, and high activity. Its performance is as follows:
Table 3: Performance of mRNA Vaccinia Capping Enzyme GMP-grade
|
Performance |
Details |
|
Source |
E. coli with vaccinia virus capping enzyme gene |
|
Optimum Temperature |
37℃ |
|
Host nucleic acid residue |
<10 fg/U |
|
Host protein residue |
<50 ppm |
|
Pathogen(HBV/ HCV/HIV) |
Negative |
|
Endotoxin |
<0.05 EU/1000 U |
|
Mycoplasma detection |
Negative |
|
Sterility |
Negative |
|
Storage Buffer |
20 mM Tris-HCl pH 8.0,100 mM NaCl,1 mM DTT,0.1 mM EDTA,0.1% Triton X-100,50% glycerin |
|
Unit Definition |
1 unit: The amount of enzyme required to incorporate 10 pmol GTP (α- 32P) into a transcript with 80 nucleotides (80 nt) at 37℃ within 1 hour. |
Cap analogs are co-transcriptional capping reagents for in vitro transcription of 5'-capped mRNAs, resulting in Cap structures that result in mRNAs with higher in vivo activity and translation efficiency.
4.4. Additional products for mRNA IVT and modification
Table 4: Additional products for mRNA IVT and modification
|
Procedures |
Product Name |
SKU |
Specifications |
|
IVT |
10623ES |
50/100 T |
|
|
10624ES |
2.5/5/25/50 KU |
||
|
10611ES |
0.5/2/10 KU |
||
|
10620ES |
10/100/1000 U |
||
|
Murine RNase inhibitor GMP-grade |
10621ES |
10/20/100 KU |
|
|
10133ES |
1 Set (4 vial) |
||
|
mRNA modification |
10614ES |
2/10/20/100 KU |
|
|
10612ES |
2/10/50/250 KU |
||
|
10619ES |
0.5/25/50/500 mL |
||
|
10132ES |
1 mL |
Regarding reading:
Yeasen Biotechnology GMP Grade mRNA In Vitro Synthesis Raw Materials
DNase I and Their Applications in Biomedicine
Murine RNase Inhibitors-Successfully eliminate RNase contamination and preserve RNA
References:
[1] Jackson, Nicholas AC, et al. "The promise of mRNA vaccines: a biotech and industrial perspective." npj Vaccines 5.1 (2020): 1-6.
[2] Maruggi, G., et al., mRNA as a Transformative Technology for Vaccine Development to Control Infectious Diseases. Mol Ther, 2019. 27(4): p. 757-772.