2×Hieff™ PCR Master Mix (With Dye)
Description
2×HieffTM PCR Master Mix contains HieffTM Taq DNA Polymerase (Cat#10101), dNTPs, and other PCR-required components. The Master Mix is stable for 3 months at 4℃ with our customized stabilizers. The pre-mix solution is optimized for conventional PCR and ready to use by adding DNA template and primers. The PCR products can be loaded directly for electrophoresis with pre-loaded bromophenol blue dye. The amplified products contain 3 '-dA protrusion and can be easily cloned into T vector. The 2×HieffTM PCR Master Mix simplifies PCR procedure and reduces contamination.
Features
- Convenient, ready-to-use mix
- Thermostable—half-life is more than 40 min at 95℃
- Generates PCR products with 3'-dA overhangs
Applications
- Routine PCR amplification of DNA fragments
- High throughput PCR
- DNA labeling
Specifications
|
Fidelity (vs. Taq) |
1× |
|
Hot Start |
No |
|
Overhang |
3 '-A |
|
Polymerase |
Taq DNA Polymerase |
|
Reaction Format |
SuperMix or Master Mix |
|
Reaction Speed |
Standard |
|
Product Type |
PCR Master Mix (2x) |
Storage
The 2×HieffTM PCR Master Mix products should be stored at -25~-15℃ for 2 years.
Figures
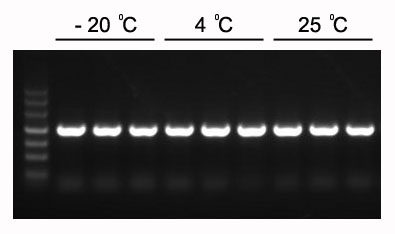
Figure 1 The expected 1.2 kb PCR products can be amplified with 2×HieffTM PCR Master Mix.
The Master Mix was stored at -20℃ for 1 year following another 3 months at 4℃ and 1 month at 25℃. Template: Arabidopsis genome. Annealing temperature: 60℃. Extension time: 40 sec.
Cited from "Ultrasensitive sensors reveal the spatiotemporal landscape of lactate metabolism in physiology and disease" Cell Metab . 2023 Jan 3;35(1):200-211.e9. doi: 10.1016/j.cmet.2022.10.002
Ver.EN20230901
[1] Wu LY, Shang GD, Wang FX, et al. Dynamic chromatin state profiling reveals regulatory roles of auxin and cytokinin in shoot regeneration. Dev Cell. 2022;57(4):526-542.e7. doi:10.1016/j.devcel.2021.12.019(IF:12.270)
[2] Li Y, Wang C, Zhang L, Chen B, Mo Y, Zhang J. Claudin-5a is essential for the functional formation of both zebrafish blood-brain barrier and blood-cerebrospinal fluid barrier. Fluids Barriers CNS. 2022;19(1):40. Published 2022 Jun 3. doi:10.1186/s12987-022-00337-9(IF:7.662)
[3] Luo T, Wang Y, Tang H, et al. An AAV-Based NF-κB-Targeting Gene Therapy (rAAV-DMP-miR533) to Inflammatory Diseases. J Inflamm Res. 2022;15:3447-3466. Published 2022 Jun 14. doi:10.2147/JIR.S362732(IF:6.922)
[4] Chen Z, Qi Z, He D, et al. Strategy for Scanning Peptide-Coding Circular RNAs in Colorectal Cancer Based on Bioinformatics Analysis and Experimental Assays. Front Cell Dev Biol. 2022;9:815895. Published 2022 Feb 25. doi:10.3389/fcell.2021.815895(IF:6.684)
[5] Yu Y, Fang L. CircRPAP2 regulates the alternative splicing of PTK2 by binding to SRSF1 in breast cancer. Cell Death Discov. 2022;8(1):152. Published 2022 Apr 2. doi:10.1038/s41420-022-00965-y(IF:5.241)
[6] Zhang J, Liu W, Li G, et al. BCAS2 is involved in alternative splicing and mouse oocyte development. FASEB J. 2022;36(2):e22128. doi:10.1096/fj.202101279R(IF:5.192)
[7] Xiao W, Li J, Zhang Y, et al. A fungal Bipolaris bicolor strain as a potential bioherbicide for goosegrass (Eleusine indica) control. Pest Manag Sci. 2022;78(3):1251-1264. doi:10.1002/ps.6742(IF:4.845)
[8] Zhang Y, Yu R, Tang J, et al. Three cytochrome P450 CYP4 family genes regulated by the CncC signaling pathway mediate phytochemical susceptibility in the red flour beetle, Tribolium castaneum. Pest Manag Sci. 2022;78(8):3508-3518. doi:10.1002/ps.6991(IF:4.845)
[9] Zhang X, Yang S, Chen W, et al. Circular RNA circYPEL2: A Novel Biomarker in Cervical Cancer. Genes (Basel). 2021;13(1):38. Published 2021 Dec 23. doi:10.3390/genes13010038(IF:4.096)
[10] Gu K, Qian D, Qin H, et al. A novel mutation in KCNH2 yields loss-of-function of hERG potassium channel in long QT syndrome 2. Pflugers Arch. 2021;473(2):219-229. doi:10.1007/s00424-021-02518-1(IF:3.657)
[11] Yang Y, Chu X, Nie M, et al. A novel long-range deletion spanning STX16 and NPEPL1 causing imprinting defects of the GNAS locus discovered in a patient with autosomal-dominant pseudohypoparathyroidism type 1B. Endocrine. 2020;69(1):212-219. doi:10.1007/s12020-020-02304-6(IF:3.235)
[12] Ullah H, Arbab S, Khan MIU, et al. Circulating cell-free mitochondrial DNA fragment: A possible marker for early detection of Schistosoma japonicum. Infect Genet Evol. 2021;88:104683. doi:10.1016/j.meegid.2020.104683(IF:2.773)
[13] Xu L, Chen Y, Shen T, Lin C, Zhang B. Genetic Analysis of PICK1 Gene in Alzheimer's Disease: A Study for Finding a New Gene Target. Front Neurol. 2019;9:1169. Published 2019 Jan 9. doi:10.3389/fneur.2018.01169(IF:2.635)
[14] Ullah H, Qadeer A, Giri BR. Detection of circulating cell-free DNA to diagnose Schistosoma japonicum infection. Acta Trop. 2020;211:105604. doi:10.1016/j.actatropica.2020.105604(IF:2.555)
[15] Yan ZC, Hua HQ, Qi GY, Li YX. Early Detection and Identification of Parasitoid Wasps Trichogramma Westwood (Hymenoptera: Trichogrammatidae) in Their Host Eggs Using Polymerase Chain Reaction-Restriction Fragment Length Polymorphism [published online ahead of print, 2022 Jun 24]. J Econ Entomol. 2022;toac095. doi:10.1093/jee/toac095(IF:2.381)
[16] Zhou Z, Yin H, Suye S, Zhu F, Cai H, Fu C. The changes of DNA double-strand breaks and DNA repair during ovarian reserve formation in mice. Reprod Biol. 2022;22(1):100603. doi:10.1016/j.repbio.2022.100603(IF:2.376)
Catalog No.:*
Name*
phone Number:*
Lot:*
Email*
Country:*
Company/Institute:*

