Library Quantification Kit for Illumina, qPCR Master Mix
Product Description
This product is a kit for the quantification of a high-throughput sequencing library on the Illumina platform. It provides DNA standards, qPCR Master Mix, dilution buffer, quantitative amplification primers, and reference dye ROX required for quantification. Among them, the DNA standards contain six 450 bp double-stranded DNA fragments, with a concentration of 20 pM-0.0002 pM. The amplification primers are designed according to the NGS library adapter sequences P5 and P7, which can ensure specific amplification of the library with complete adapters. The qPCR Master Mix is a Dye-Based hot-start qPCR master mix, which can efficiently and specifically, amplifies samples with different lengths, and different GC or AT contents, and achieve accurate quantification.
Product Application
This product is designed to accurately quantify libraries prepared for high-throughput sequencing on the Illumina platform. It is suitable for quantifying libraries prepared in any way that matches the NGS library adapter sequences P5 and P7, and the concentration is not less than 0.0002 pM. In addition to NGS library quantification, the kit can also be used to detect the degree of library contamination in the experimental environment during the preparation of the Illumina library.
Product Component
|
Components |
Volume |
|||
|
12302-A |
Hieff NGSTM SYBR qPCR Master Mix (2×) |
5×1 mL |
- |
- |
|
12302-B |
qPCR Primer Mix |
2×0.5 mL |
- |
- |
|
12302-C |
50×Low Rox |
200 μL |
|
- |
|
12302-D |
50×High Rox |
200 μL |
- |
- |
|
12303 |
Hieff NGS™ Library Dilution Buffer |
- |
50 mL |
- |
|
12307 |
Hieff NGS™ Library Quantification Kit for Illumina, DNA Standard1-6 |
- |
- |
6×96 μL |
Instrument Model
|
Category |
Instrument Model |
|
No ROX added |
Bio-Rad CFX96™, CFX384™, iCycler iQ™, iQ™5, MyiQ™, MiniOpticon™, Opticon®, Opticon 2, Chromo4™; Cepheid SmartCycler®; Eppendorf Mastercycler®ep realplex, realplex 2 s; Illumina Eco qPCR; Qiagen/Corbett Rotor-Gene®Q, Rotor-Gene®3000, Rotor-Gene®6000; Roche Applied Science LightCyclerTM480; Thermo Scientific PikoReal Cycler |
|
Add 50×High Rox |
Applied Biosystems 5700, 7000, 7300, 7700, 7900, 7900HT, 7900HT Fast; StepOne™, StepOnePlus™. |
|
Add 50×Low Rox |
Applied Biosystems 7500, 7500 Fast, ViiA™7; Stratagene MX4000™, MX3005P™, MX3000P™. |
[Note]: Select the appropriate reference dye ROX according to the type of instrument.
Shipping and Storage
All the components are shipped with an ice pack and should be stored at -20°C. The qPCR Master Mix and ROX can be stored in dark for 12 months, unless used repeatedly in a short period, in which case the qPCR Master Mix can be thawed and temporarily stored in the dark at 4°C for 3 months.
Cautions
I. About operation
1. For your safety and health, please wear lab coats and disposable gloves for the operation.
2. It is recommended to physically separate the sample preparation area, reaction system preparation area, and sample addition area.
3. It is recommended to perform all operations on ice boxes before performing qPCR testing on an instrument.
4. To ensure product quality, avoid repeated freezing and thawing more than 30 times.
5. The sensitivity of qPCR technology is high, and accurate pipetting is crucial to the reliability of the detection results. Accuracy of operation can be guaranteed by paying attention to the following details:
a - Make sure that all reagents and samples have been completely thawed and mixed thoroughly before the experiment, and the liquid is collected to the bottom of the tube after brief centrifugation.
b - Use a pipette tip with a filter element as much as possible to avoid aerosol pollution;
c - Avoid using Multichannel Pipette;
d - The pipette tip should be replaced every time to avoid cross-contamination;
e - Avoid the pipette tip going deep into the liquid when sucking the liquid, so as to prevent the liquid from sticking to the outside of the pipette head;
f - When discharging liquid, the pipette tip should be as close to the bottom of the reaction tube as possible;
g - After discharging the liquid, pipette up and down 2-3 times to rinse the pipette tip;
h - After discharging the liquid, check that no liquid remains at the tip of the pipette tip.
6. For research use only!
II. About library dilution
The library needs to be diluted to the effective Ct range of the standard curve for quantification. The dilution ratio can refer to past experience or the concentration determined by other assay methods as a reference (such as NanoDrop®, Qubit® or Bioanalyzer). Note that high-concentration DNA solutions are viscous and have poor dispersion of DNA molecules. Serial dilutions are recommended during the dilution process (for example, two serial 1:100 dilutions are performed instead of only one 1:10000 dilution). The range of library concentrations that can be quantified using this kit is shown in the table below.
Since DNA is easily degraded in a non-buffered environment, it is recommended to use dilution buffer (10 mM Tris-HCl, pH 8.0 [25°C]; 0.05% Tween 20) (cat#12303ES50) to dilute the library to ensure the quality of the assay samples. Never use water as a diluent. When assaying, the library should be diluted and stored on ice. Do not use the diluted library prepared and stored in the past. If the sample needs to be re-assayed, a freshly diluted sample will need to be prepared for testing.
|
DNA Standard |
Molarity |
Concentration |
Copy Number Concentration |
|
STD 1 |
20 pM |
5.5 pg/μL |
12×10 6 copies/μL |
|
STD 2 |
2 pM |
0.55 pg/μL |
12×10 5 copies/μL |
|
STD 3 |
0.2 pM |
0.055 pg/μL |
12×10 4 copies/μL |
|
STD 4 |
0.02 pM |
0.0055 pg/μL |
12×10 3 copies/μL |
|
STD 5 |
0.002 pM |
0.00055 pg/μL |
12×10 2 copies/μL |
|
STD 6 |
0.0002 pM |
0.000055 pg/μL |
12×10 1 copies/μL |
III. Contamination and No-template Controls
1. When performing qPCR experiments, good experimental practices should be ensured to prevent contamination of the work area, reagents, consumables, instruments, DNA standards, etc. It is recommended to physically separate the reaction system preparation area from the template preparation area, and regularly use 0.5% sodium hypochlorite or 10% bleach for each experiment.
2. During the preparation of the reaction system, DNA Standards should be added in the order from low concentration to high concentration (DNA Standard 6 to 1), and new pipette tips should be replaced with each pipetting to avoid aerosol contamination.
3. The NTC negative control reaction should be performed in parallel for each experiment, and the amplification specificity and system contamination degree analysis should be carried out in conjunction with the melting curve. Because the amplification primer sequences are fixed sequences designed by the Illumina® platform rather than qPCR-specific primer sequences, and the number of amplification cycles is large, it is normal to generate primer-dimer amplification resulting in Ct production for the NTC negative control reaction. Normally, Ct (NTC) > Ct (DNA Standard 6) + 3.
IV. Amplification curve baseline settings
As the molarity of DNA Standard 1 is much higher than that of conventional qPCR templates, its Ct value is often very small. However, most qPCR instruments set 3-15 cycles as Baseline by default, and sometimes the Ct value of DNA Standard 1 is mistaken for the noise background during the measurement, which in turn affects the standard curve creation. Manually setting the Baseline to cycles 1-3 can effectively avoid this from happening.
V. Melting curve analysis
The melting curve is very important when performing contamination analysis with the NTC negative control and confirming the maximum effective Ct of the standard curve. It is recommended to perform the melting curve acquisition step for each experiment. All melting curves generated by the DNA Standards for this product exhibit a characteristic single peak.
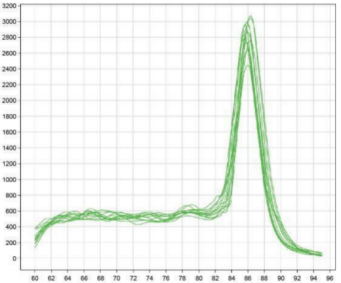
Figure 1. Library standard melting curve
Special Note
Cat#12302 and Cat#12307 are not sold separately. Must be sold in combination!
Catalog No.:*
Name*
phone Number:*
Lot:*
Email*
Country:*
Company/Institute:*