Reverse transcription, as one of the basic techniques of molecular biology research, is the process of synthesizing cDNA using RNA as a template and through reverse transcriptase. However, several parameters such as prepared RNA templates, chosen primers, reverse transcriptase, and reaction conditions may readily alter apparently straightforward reverse transcription operations. The next sections mostly cover how to choose reverse transcriptase, which is the most important enzyme raw material for reverse transcription.
1. What is Reverse transcriptase?
2. What are types of reverse transcriptase?
3. Reverse transcriptase from the Yeasen Hifair™ series
4. Performance demonstration of Hifair™ III Reverse Transcriptase
5. Reverse Transcription Products are available for purchase
1. What is Reverse transcriptase?
Reverse transcriptase is a kind of DNA polymerase that performs three different functions: RNA-directed DNA polymerase activity, RNase H activity, and DNA-directed DNA polymerase activity.
1. Synthesize the initial cDNA strand complementary to the template RNA, resulting in an RNA-DNA hybrid strand, using RNA-guided DNA polymerase activity.
2. RNase H activity hydrolyzes the RNA strand in the cDNA-RNA hybrid strand to produce single-stranded cDNA.
Note: Because RNase H may destroy the RNA template before full-length reverse transcription is accomplished, lowering or eliminating RNase H activity in reverse transcriptase improves the capacity of reverse transcriptase to create long-chain cDNA.
3. Catalyzes the synthesis of a complementary second cDNA strand from single-stranded cDNA using DNA-directed DNA polymerase activity.
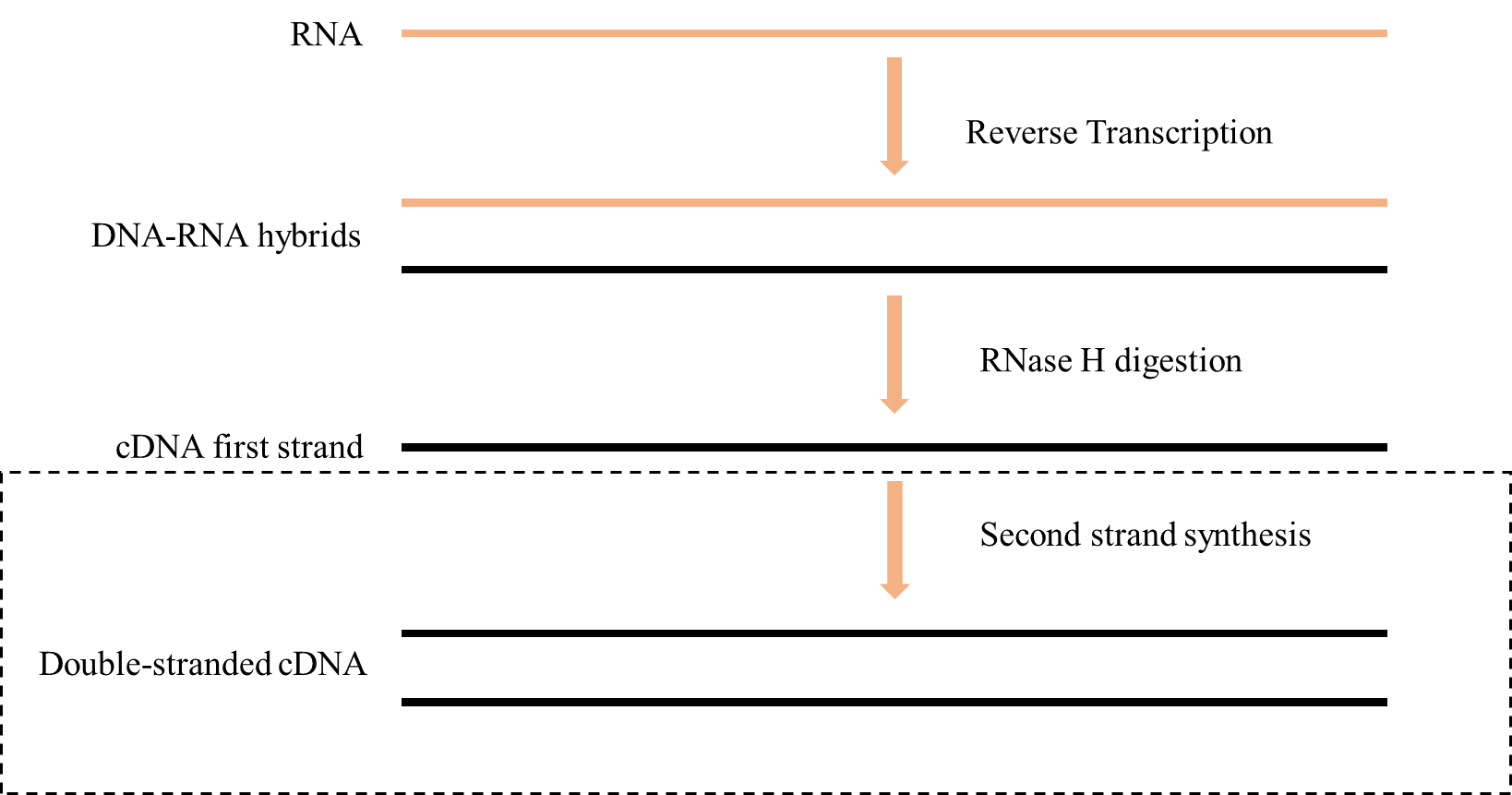
Figure 1. Schematic diagram of the reverse transcription process
Reverse transcription consists of different structural enzymes with different biochemical activities. There are 6 things to note about the nature of reverse transcriptase. They are DNA polymerase activity, RNase H activity, thermostability, processivity, fidelity, and terminal nucleotidyl transferase (TdT) activity.
RNase H can simultaneously cleave RNA templates in RNA:cDNA hybrid strands during synthesis. Since the RNA template may be degraded before full-length reverse transcription is completed, RNase H enzymatic activity is a factor that needs to be avoided during the synthesis of long cDNA fragments. And because RNase H competes with the active polymerase in reverse transcriptase, RNase H enzymatic activity can reduce reverse transcription efficiency.
Factors affecting cDNA synthesis in reverse transcriptase include the ability to withstand high temperatures. Elevating the reaction temperature can denature RNAs with robust secondary structure and/or high GC content, allowing reverse transcriptase to read the sequence. Therefore, at the higher reaction temperature, reverse transcription can achieve full-length cDNA synthesis with higher yield and better reverse transcription of RNA into cDNA.
The synthetic capacity of reverse transcription refers to the number of nucleotides incorporated into a single binding site of an enzyme. Therefore, a reverse transcriptase with high synthesis capacity can synthesize longer cDNA strands in a shorter reaction time.
The fidelity of reverse transcription refers to the sequence accuracy during the reverse transcription of RNA to DNA. Fidelity and reverse transcription errors are inversely proportional.
Reverse transcriptases exhibit TdT activity only when the reverse transcriptase contacts the 5' end of the RNA template. At this point, it adds 1 to 3 additional nucleotides to the cDNA end and exhibits specificity for double-stranded nucleic acid substrates.
By developing and modulating the above intrinsic properties of reverse transcriptase, successful cDNA experiments can be performed. Not only can it be used for research its use, but also an important tool for other applications in the field of molecular biology.
2. What are types of reverse transcriptase?
Different reverse transcriptases have different functional activities and qualities, which impact the length, yield, and capacity to reverse complicated and challenging RNA templates, among other things. Most commercially available reverse transcriptases are derived from the avian ameloblastoma virus (AMV) or the Moloney murine leukemia virus (M-MLV). RNase H activity, reverse transcription temperature, duration, and yield vary greatly, yet each has benefits and drawbacks.
Through genetic modification based on M-MLV, Yeasen has produced Hifair™ series reverse transcriptases with reduced RNase H activity, improved thermal stability, and stronger continuous synthesis capacity to further increase reverse transcription efficiency. The features of many kinds of reverse transcriptases are listed in Table 1.
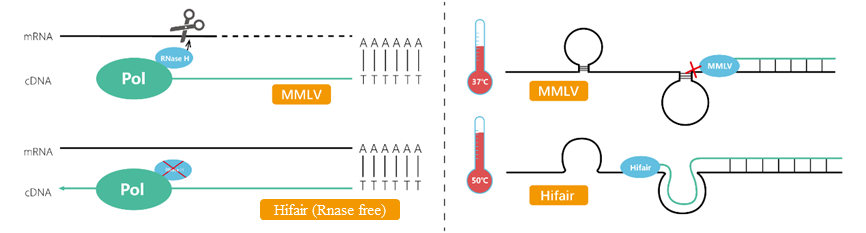
Figure 2. Schematic of reverse transcriptase
Table 1. Comparing reverse transcriptases
|
AMV Reverse Transcriptase |
M-MLV Reverse Transcriptase |
Yeasen Hifair™ Reverse Transcriptase |
|
|
Temperature |
42℃ |
42-60℃ |
42-60℃ |
|
Reaction time |
60 min |
30-60 min |
15-30 min |
|
Yield |
Middle |
Low |
High |
|
Length |
Short |
Middle |
Long,up to 19.8kb |
|
RNase H activity |
High |
Middle |
Low |
3. Reverse transcriptase from the Yeasen Hifair™ series
Yeasen now offers the Hifair™ family of reverse transcriptases in a range of dosage forms, including single enzyme, premix, and kit formats, which you may choose based on your specific requirements. The features and suggested uses of Yeasen various reverse transcriptases are shown in Table 2.
Table 2. Guide to choosing a reverse transcriptase
|
Product name |
Cat# |
Features |
Application Suggestions |
|
HifairTM Ⅲ Reverse Transcriptase (Inquire) |
11111ES |
Long synthesis duration, thermally stable (up to 60°C), and suited for complicated templates. |
RT-Lamp,Two steps RT-PCR/qPCR,Clone. |
|
HifairTM IV Reverse Transcriptase (Inquire) |
11112ES |
Lack of RNase H activity, high yield, and good continuous synthesis ability. |
Construction and sequencing of an RNA library. |
|
11300ES |
Exceptional compatibility. |
One step RT-PCR/qPCR. |
A reverse transcriptase utilized in RT-Lamp isothermal amplification is discussed in the following: Reverse Transcriptase Hifair™ III The enzyme has a good thermal stability and a rapid production speed. It can survive reactions up to 60°C, making it ideal for reverse transcription of RNA templates with complicated secondary structures. The enzyme also lacks RNase H activity and increases its elongation ability, with a synthetic length of 19.8 kb. Furthermore, the enzyme has a higher affinity for the template, making it appropriate for reverse transcription of low-copy genes and tiny template quantities.
4. Performance demonstration of Hifair™ III Reverse Transcriptase
4.1 Resistant to 60°C reaction temperature, suitable for reverse transcription of complex RNA templates.
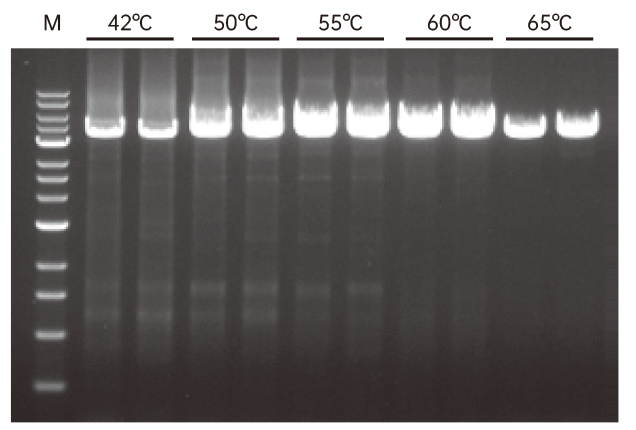
Figure 3. Reverse transcription was done at 42°C-65°C using 500 ng of total RNA from 293T cells as a template and Hifair™ III reverse transcriptase (Cat#11111ES). Take 1 μL of cDNA as a template, and use Hieff Canace™ Gold High Fidelity DNA Polymerase (Cat#10148ES)(Inquire) to amplify the TFRC gene (4.4 kb). M:1 kb DNA ladder.
4.2 Without RNase H activity, 19.8 kb cDNA can be produced
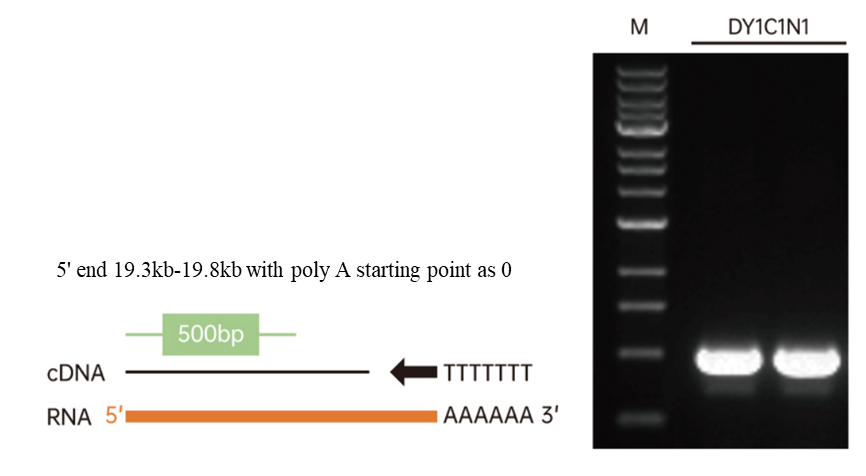
Figure 4. cDNA was generated using Hifair™ III reverse transcriptase (Cat#11111ES) and 500 ng of total RNA from 293T cells as template and oligo dT as primer. Take 1 μL of cDNA as template, and use Hieff Canace™ Gold High Fidelity DNA Polymerase (Cat#10148ES)(Inquire) to amplify 500 bp of the 5' of DY1C1N1 gene (19.8 kb). M: 1 kb DNA ladder.
4.3 Suitable for reverse transcription of genes with different GC contents and different expression abundances
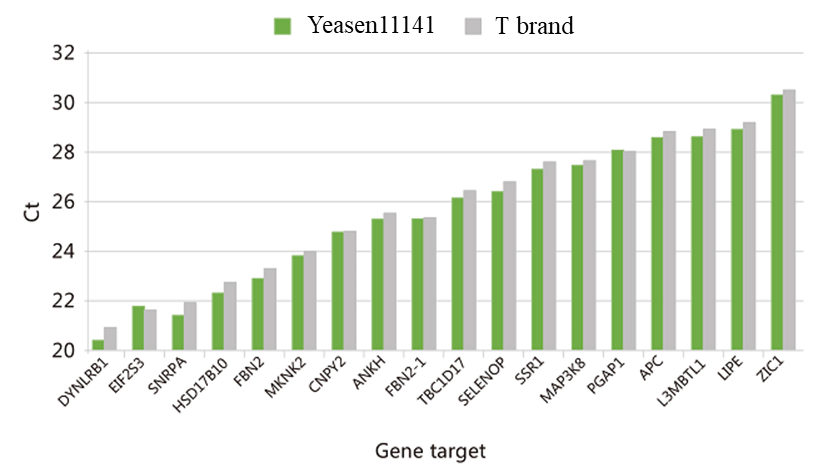
Figure 5. Using 300 ng of total RNA from 293T cells as a template, cDNA was synthesized using Hifair™ Ⅲ 1st Strand cDNA Synthesis SuperMix for qPCR (Cat#11141ES), T brand. Take 1 μL of cDNA as template, and use Hieff UNICON™ Power qPCR SYBR Green Master Mix (Cat#11195ES)(Inquire) to amplify 20 genes with different GC content (25-65%) and different expression abundances.
5. Reverse Transcription Products are available for purchase
Reverse transcription products provided by Yeasen are as follow:
Table 3. Reverse transcription products provided by Yeasen
|
Product positioning |
Product name |
Cat# |
|
Enzyme |
HifairTM III Reverse Transcriptase (200 U/μL) (Inquire) |
11111ES |
|
HifairTM IV Reverse Transcriptase (200 U/μL) (Inquire) |
11112ES |
|
|
11300ES |
||
|
One-step gDNA removal + Reverse transcription master mix |
11142ES |
|
|
Reverse transcription master mix |
HifairTM Ⅲ 1st Strand cDNA Synthesis SuperMix for qPCR (gDNA digester plus) |
11141ES |
|
Reverse transcription kit |
HifairTM III 1st Strand cDNA Synthesis Kit (gDNA digester plus) |
11139ES |
6. Ordering products
The other products provided by Yeasen are as follow:
Table 4: The other products provided by Yeasen
|
Product positioning |
Product name |
Cat# |
|
Hot start Taq polymerase |
10726ES |
|
|
Double-Block anti-Taq DNA Polymerase antibody |
31303ES |
|
|
Isothermal amplification of Bst polymerase |
14402ES |
|
|
Murine RNase inhibitor |
10603ES |
|
|
UDG |
Uracil DNA Glycosylase (UDG/UNG), heat-labile (1 U/μL) (Inquire) |
14455ES |
|
High purity nucleotides |
10128ES |
|
|
qPCR master mix |
11184ES |
|
|
qPCR master mix |
Hieff UNICONTM TaqMan Multiplex qPCR Master Mix (UDG plus) (inquire) |
13171ES |
|
RT-qPCR master mix |
Hieff UniconTM C58P1 nCov Multiplex One Step RT-qPCR Probe Kit (inquire) |
13173ES |
Note: All of the above enzymes are available in high-concentration or glycerol-free versions.
7. Regarding Reading
Deep learning the core enzyme raw materials used in the nucleic acid detection of SARS-CoV-2
COVID-19 nucleic acid detection raw material solution: fast, freeze-dried
References:
[1] Liu C X, Li X, Nan F, et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity[J]. Cell, 2019, 177(4): 865-880. e21.(IF31.398)
[2] Fan H, Hong B, Luo Y, et al. The effect of whey protein on viral infection and replication of SARS-CoV-2 and pangolin coronavirus in vitro[J]. Signal transduction and targeted therapy, 2020, 5(1): 1-3. (IF13.493)
[3] Wang J., et al., The mycobacterial phosphatase PtpA regulates the expression of host genes and promotes cell proliferation[J]. Nat Commun. 2017 Aug 15;8(1):244.(IF 12.353)
[4] Zhou L, Hou B, Wang D, et al. Engineering Polymeric Prodrug Nanoplatform for Vaccinatio Immunotherapy of Cancer[J]. Nano Letters, 2020. (IF12.279)
[5] Xu L, Xu S, Sun L, et al. Synergistic action of the gut microbiota in environmental RNA interference in a leaf beetle[J]. Microbiome, 2021, 9(1). (IF 11.607)
[6] Arezi B, Hogrefe HH. Escherichia coli DNA polymerase III epsilon subunit increases Moloney murine leukemia virus reverse transcriptase fidelity and accuracy of RT-PCR procedures. Anal Biochem. 2007 Jan 1;360(1):84-91. (IF 3.191)